Sticklebacks, selection shadows, and salinity shifts: phenotypic plasticity in novel environments
1 May 2017
Phenotypic plasticity is the ability of one genotype to produce multiple phenotypes depending on external environmental conditions (see our slideshow for further information). It comes in many forms: from permanent developmental shifts in early life to ongoing, reversible behavioural responses to fluctuating environmental conditions. Although widespread, its role and importance in evolutionary dynamics is not yet fully understood.
Much of the research on phenotypic plasticity is looking at it on an individual level: the evolutionary consequences of an individual being able to adapt to the environment it is in. My research instead focuses on the population-level effects of phenotypic plasticity. Since not every individual (or genotype) is plastic in the same way, the same population may react differently to different environments. In some environments, all individuals in a population may be phenotypically very similar – in others, individuals from the same population may show large phenotypic differences. This has implications for the evolutionary potential of that population, based on the environment it is in.
The idea that populations can show different patterns of phenotypic variation based on the environment is actually fairly obvious when you consider the nature of genotype x environment (G x E) interaction. What I’m looking for is patterns in this phenomenon. Specifically, I’m interested in the potential for plastic traits in a population to show greater phenotypic variation in novel environments.
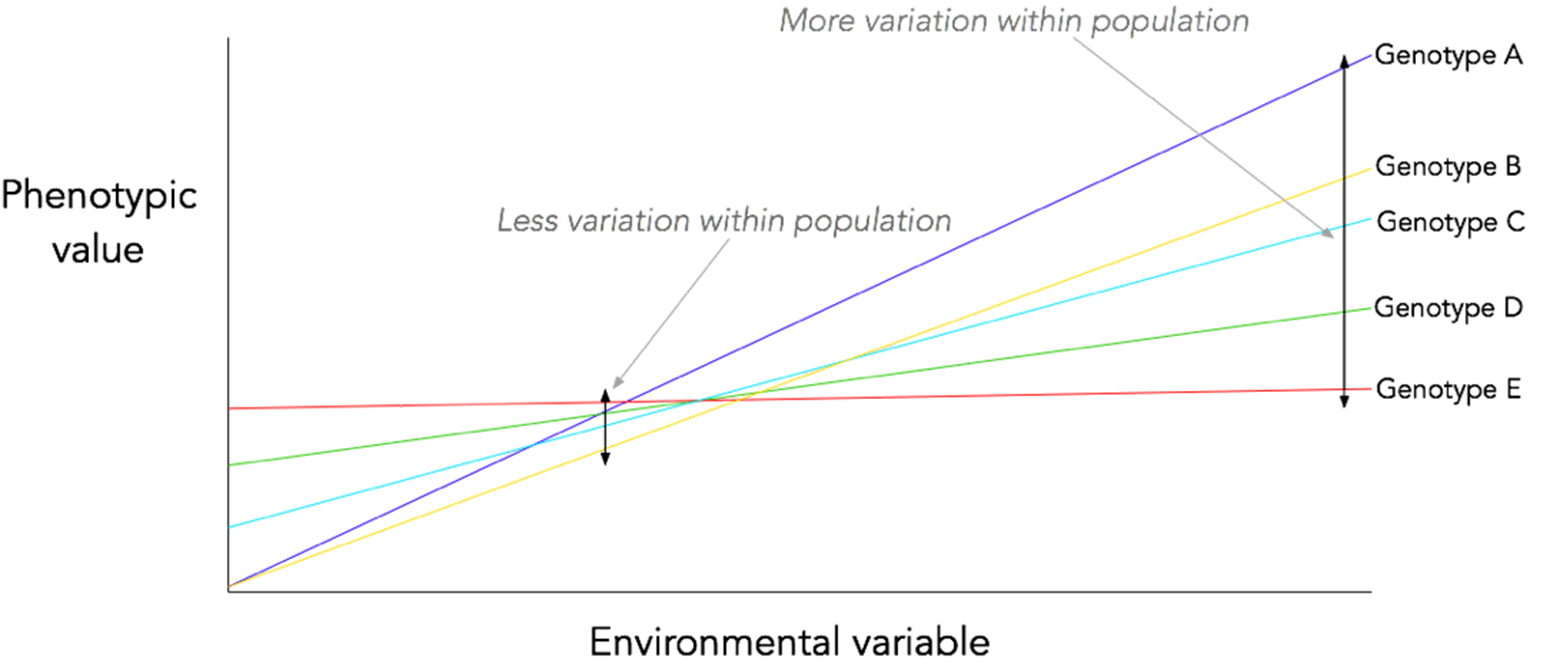
Theoretically, there is reason to believe that this may be the case. Phenotypic plasticity often relies on different gene expression patterns based on the environment. The expression patterns of genes in commonly-encountered environments are under selection (and, in the case of stabilising selection, will show reduced phenotypic variation as a result) – but their expression patterns in rarely-encountered environments are not, meaning that any mutation which only affects the gene expression in the rarely-encountered environment will not be removed by selection and can spread by drift. This concept is in fact well-known in the field of senescence research1,2: it is a central tenet of many of the evolutionary theories of senescence i.e. that the ‘selection shadow’ caused by extrinsic mortality means that mutations which are only deleterious in old age (i.e. in a small percentage of the population) are not strongly selected against.
This is the idea associated with the spread of ‘cryptic genetic variation’ – genetic variation which only turns into phenotypic variation in certain conditions (usually environmental, but the ‘condition’ can include the genomic background as well). It has been hypothesised to play an important role in adaptation to new environments and evolutionary innovations – but many questions about its possible role in evolutionary dynamics remain. How common is it? Are some traits or mechanisms of plasticity more likely to result in cryptic genetic variation than others? Is there a bias in the fitness of the cryptic genetic variants, or is it random? And – perhaps most importantly – are there patterns in the type of environment required to reveal cryptic genetic variation?

My research aims to look for these patterns, using salinity-based plasticity of the three-spined stickleback (Gasterosteus aculeatus). These tough little fish are able to cope with both sea- and fresh-water habitats, using a combination of neat physiological and behavioural tricks. Some of them, however, have radiated into freshwater habitats and stayed there permanently, creating a useful natural experiment. The freshwater ones are descended from anadromous ancestors, but are no longer exposed to high-salinity environments. By comparing the response to a range of salinities of fish from different populations (both anadromous and freshwater) I can investigate what happens to the salinity-based plasticity of stickleback traits when they are no longer stabilised by selection for seawater tolerance.
This will allow me to see whether cryptic genetic variation (released in high-saline environments) has begun to creep in for traits in the freshwater fish. Since there are multiple stickleback traits which are plastic in response to salinity, I will also be able to look at whether the patterns are different between traits. The repeated invasions into freshwater habitats mean that I can compare different populations which have undergone similar environmental shifts – and even get an idea of the timescale of any changes I see, since different populations invaded freshwater environments at different times. Most importantly, I will be looking for patterns in the population-level variation in traits. Will my freshwater fish show higher levels of trait variation in saline environments than in freshwater, relative to my anadromous fish?
This last question is key: it is the most revealing about the evolutionary dynamics of phenotypic plasticity. If plastic traits do show predictable patterns of population-level variation based on the environment, it is an argument in favour of explicitly considering trait plasticity in evolutionary dynamics. If not, it may be that there is no need to make plastic traits an evolutionary ‘special case’.
This PhD research project is part of ‘An experimental test of plasticity-led evolution’ (Project #14 in the EES research program) under the theme ‘Evolutionary Diversification’. Please don’t hesitate to get in touch with [email address=”hcsj@st-andrews.ac.uk” name=”Helen”] for discussion and collaboration.
Project leader: Susan Foster (Clark University)
Project supervisors: Kevin Laland, Mike Webster (University of St Andrews)
1. Medawar PB 1952. HK Lewis & Co London.
2. Williams GC 1957. Evolution 11:398-411.