Research project 9: The evolution of inclusive heredity through the genomic interactions of symbionts
20 February 2017
‘The evolution of inclusive heredity through the genomic interactions of symbionts’ is one of five research projects under the third theme, Inclusive Inheritance, of the Extended Evolutionary Synthesis research program. Michael Wade and Armin Moczek (both at Indiana University), are using theoretical models and experiments in dung beetles to investigate how the evolution of symbiotic host-microbiota interactions shape heredity.
During each of the major evolutionary transitions (see also Research project 7: Origins of organismal complexity) the way that information is transmitted from one generation to the next has changed1. For example, during evolution of eukaryotic cells from prokaryotic cells, a complex system of epigenetic marks (chemical modifications to DNA or associated proteins) arose as a mechanism of transmitting environmental information to daughter cells. A second example is found in the transition from single-celled to multi-celled organisms, where separation of the germline uncoupled reproduction of cells from reproduction of the organism, thereby reorganizing the way genetic information is transmitted. The evolution of heredity in complex organisms, like humans, involves the coordinated transmission of genes but also of environments, including communities of microbes (the microbiome).
Organisms and their microbiomes participate in a symbiotic (mutually beneficial) relationship, sometimes referred to as the holobiont. ‘Genomic conflict theory’ predicts that organism and symbiont coexistence should break down due to ‘cheater genotypes’ – that is, individuals that behave ‘selfishly’ at a cost to other individuals2,3. In spite of this, holobionts are remarkably stable and some researchers view this as a difficultly for traditional evolutionary explanations4.
The Extended Evolutionary Synthesis’ view of heredity as reconstruction of life cycles, rather than transmission of genes, provides insights into the solution to the holobiont dilemma. This perspective emphasizes that stable recurrence of holobionts is the result of both natural selection and inheritance of the environmental niches that organisms construct for each other. This shared ecological context can provide the starting point for more elaborate and intimate relationships that characterize the holobiont. Indeed, during the evolution of the holobiont, inheritance of microorganisms evolved from horizontal (contagious infection) to vertical (from parents to offspring).
Investigators Wade and Moczek are studying how transmission patterns of environmental information and genomic information (from maternal, zygotic and microbial genomes) arise, coevolve, respond and feedback upon one another. They are taking complementary theoretical and empirical approaches.
For the theoretical work, Wade and Moczek are determining the necessary theoretical conditions for an evolutionary transition from a free-living, contagiously (or horizontally) inherited, ancestral symbiont to a vertically transmitted, derived mutualist. They are also developing theoretical models that will establish if the signature of niche construction between host and symbionts can be seen in the interdependence and arrangement of the genomes that together constitute the holobiont. This allows us to predict how genomic interactions might act through mutual niche construction to affect the evolution of genetic architecture.
By extending quantitative genetic models, the researchers will test the fitness consequences of microbiomes with many mutualistic species. If large microbiomes result in greater stability, it would illustrate a fundamental difference between genomic conflict and niche construction theory. Classic ecological theory predicts that community stability should decline as species number increases in competing communities5, which also holds true for genomic conflict theory. In contrast, niche construction theory predicts that community stability may increase with increasing species numbers for mutualistic communities, because of the indirect interactions that arise through niche construction6.
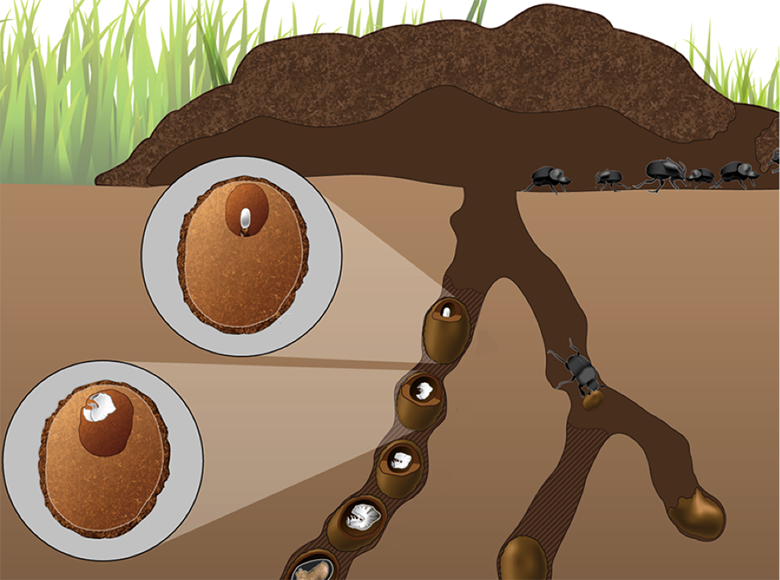
Predictions generated by the theoretical models above, concerning fidelity of transmission through niche construction, interdependence in development, number of symbionts, and their genomic diversity, will be tested in experiments using Onthophagus dung beetles. This model system combines rich ecological diversity with easy experimental accessibility and rapidly growing transcriptomic and genomic resources7. Onthophagus taurus, the focal species of this study, is a typical tunneling dung beetle. During reproduction, adult females provision eggs with all the food larvae need to complete development in the form of a brood ball made of dung and buried 10-40 cm below ground. Mothers also provide a microbial contribution in the form of a small ‘pedestal’ made of maternal feces onto which the egg is positioned8. Upon hatching, larvae consume this pedestal.
Wade and Moczek will experimentally manipulate the nutritional microbiome to test their theoretical predictions. They will use artificial microbiomes varying in their degree of microbial community deficiency and test whether the developmental consequence of these differentially deficient microbial communities can be predicted from theory. This will also determine whether there are groups of microbes that are essential for developmental transitions in Onthophagus beetles and how transmission of maternal symbionts affects gene diversity within and between host populations of Onthophagus taurus.
1. Maynard Smith J, Szathmary E. 1997. Oxford UP. 2. Wade MJ, Priest NK, Cruickshank T. 2009. U Chicago P. 3. Wade MJ. 2014. Cold Spring Harbor Perspectives 73-81. 4. Gilbert SF, Sapp J, Tauber AI. 2012. Quart Rev Biol 87:325-341. 5. May RM. 1973. Princeton UP. 6. Odling-Smee FJ, Laland KN, Feldman MW. 2003. Princeton UP. 7. Kijimoto T, et al. 2013. WIREs Dev Biol 2(3):405-18. 8. Estes AM, et al. 2013. PLoS One 8(11) e79061.