Conditional gene expression, developmental plasticity, and the diversification of horned beetles
21 July 2017
Developmental plasticity is the ability of an organism to adjust aspects of its phenotype in response to environmental conditions (both internal and external). Plasticity is a well-studied phenomenon, and a growing number of studies implicate developmental plasticity in facilitating and shaping adaptive evolution. We used three horned beetle species to learn more about possible relationships between developmental plasticity and species diversification, and the underlying mechanisms. Specifically, we asked whether plasticity in gene expression within a species predisposes genes to contribute disproportionately to genetic divergence among species. We find that genes that within a species exhibit plastic expression conditional upon external (nutrition) or internal (body region, sex) circumstances do indeed contribute significantly more to diversification among species, though exactly how to best interpret these findings remains challenging.
We chose to study horned beetles in the genus Onthophagus because it is an extraordinarily diverse genus (over 2000 species), and this diversity is organized primarily along three axes of phenotypic variation:
(i) horn position: horns can develop on the head, the thorax or both
(ii) sex bias in horn growth: horns usually develop only on males (or are more exaggerated on males) but in some species this sex bias is reversed and horns grow only on females
(iii) sensitivity of horn growth to nutrition: horn growth within sexes can show modest to extreme responsiveness to nutrition levels, creating enormous horns in the males of some species but not others
We selected three species of Onthophagus beetles that vary across these morphological traits: O. taurus, O. nigriventris and O. sagittarius. The figure below illustrates the anatomical position, sexual dimorphism and nutritional polyphenism of horn growth in the three species. O. taurus and O. sagittarius are the most closely related, with a common ancestor 5-10 million years ago (MYA) and they share a common ancestor with O. nigriventris approximately 30 MYA.
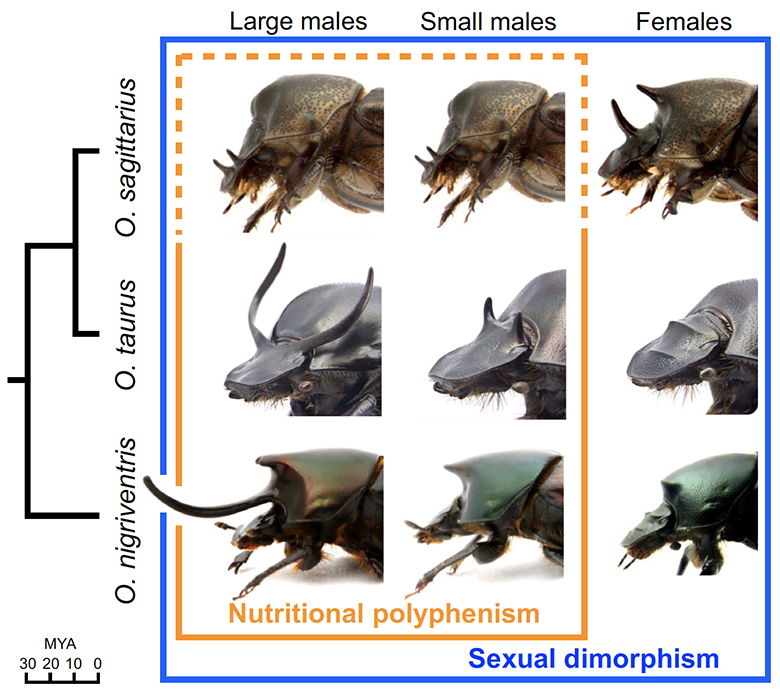
Using these three species as our microcosm, we sought to test the hypothesis that genes whose differential expression underlies the morphological differences between morphs and sexes within species are the same genes (or belong to the same functional groups) that underlie DNA sequence divergences, and thus evolved genetic changes, between species. For example, do genes expressed differentially under high or low nutrition within a species also undergo genetic changes across species that have diverged heritably in nutritional sensitivity? Is it the same group of genes that is conditionally expressed in primarily male or female beetles of one species the same group that also exhibit sequence divergences when species are compared that have diverged in degree or direction of horn growth sex bias?
The answer is yes. Take the two most closely related species, O. taurus and O. sagittarius for example. These species exhibit divergences across all three morphological axes we studied:
(i) horn position: horns grow on the head in O. taurus and on both the head and thorax in O. sagittarius; (ii) sex bias in horn growth: horns are greatly elaborated in O. taurus males but in O. sagittarius females; and (iii) horn growth sensitivity to nutrition: in O. taurus horns grow bigger on well-nourished beetles whereas horn growth in O. sagittarius is insensitive to nutrition levels.
Using gene expression data from O. taurus beetles, we produced three sets of conditionally expressed genes: body region-specific genes, sex-specific genes and nutrition-responsive genes. We tested our hypothesis by sequencing RNA from O. taurus and O. sagittarius beetles and assessing the rate of synonymous mutations (no effect on amino acid sequence) versus non-synonymous mutations (change in amino acid sequence). If our hypothesis is correct, we predicted to find greater amino acid divergence between O. taurus and O. sagittarius in all three gene sets (of conditionally expressed genes) compared with randomly selected genes. Indeed, this was the case with very high statistical significance in all three comparisons (false discovery rate (FDR) of P < 10-10).
The other species comparisons also supported our hypothesis. O. taurus and O. nigriventris differ in horn location and degree of nutrition-responsive growth and accordingly the rate of amino acid divergence was elevated in the body region-specific and nutrition-responsive gene sets. This was not the case for the sex-specific gene set, as both species exhibit the same degree and direction of sex-biased horn growth. The third species pair, O. nigriventris and O. sagittarius differ in the location of male horns, the nutrition-responsive growth of male horns and the sex that grows the most exaggerated horns. Elevated amino acid sequence divergence between the two species was found in the sex-specific and the body region-specific gene sets but surprisingly not in the nutrition-responsive gene set. Thus, only a single out of a total of nine comparisons did not conform to our predictions, providing overall strong support for our initial hypothesis.
Our results show a positive relationship between conditional gene expression within species and genetic divergence among species. This supports the idea that developmental plasticity (in the form of conditional gene expression during development) biases the evolution of morphological traits. Of course, the direction of causality could be reversed. That is, fast evolving genes could more readily be co-opted into conditional expression patterns over time, imposing constraints on the evolution of developmental plasticity. Our data do not allow us to rule out this possibility, and thus more work is clearly needed.
Taken in the context of other empirical and theoretical findings, however, we believe our data provide further support for a role for developmental plasticity in biasing the evolution of morphological traits. In this case, developmental plasticity comes in the form of conditional gene expression, i.e. gene expression influenced by the internal and/or external environment. Previous theoretical and empirical work suggests that in principle conditionally expressed genes may be able to evolve more rapidly because (i) their very specific expression reduces the probability that sequence changes may have detrimental effects elsewhere (reduced pleiotropic constraint) or (ii) because at least for genes that are biased toward one sex or nutritional conditions, not all gene copies within a population are expressed every generation, resulting in a relaxation of the strength of selection, and an accumulation of selectable variation able to fuel genetic differentiation. In either case, conditionally expressed genes within a species may have the potential to contribute important developmental genetic substrate to facilitate divergence among species. This could bias adaptive radiations along certain morphological axes, such as the ones we studied.
For more detail, read the paper here:
You can contact the lead author, Melissa Pespeni, via [email address=”mpespeni@uvm.edu” name=”email”].
This study is part of the EES research program project #5, entitled, The role of developmental plasticity in innovation and diversification of Onthophagus beetles, under the theme, Evolutionary innovations.